The extraordinary ability of animals to rapidly evolve in response to predators has been demonstrated via genetic sequencing of a waterflea population across nearly two decades.
In a new study published in Nature Communications, together with scientists at the Universities of Birmingham in the UK, the KU Leuven in Belgium, and the Leibniz Institute for Freshwater Ecology and Inland Fisheries, Berlin, we were able to identify more than 300 genes that vary in the genome of the waterflea, Daphnia magna.

These genes, which account for about 3 percent of all sequenced waterflea genes, underpin changes in behavioural and life history traits that improve survival when exposed to predators.
Strikingly, evolution in response to predation pressure occurs within just a few generations. It is mediated by so-called standing genetic variation – the amount of genetic diversity harboured by a given natural population. This standing genetic diversity is of crucial importance to support rapid adaptation. Reversely, reducing the genetic diversity of natural populations can reduce their ability to adapt to environmental change.
We were able to quantify the genetic diversity of one particular waterflea population over nearly two decades and show clearly how rapid evolution took place in response to environmental pressures.
The waterflea is central to the food webs of lakes and ponds. Its life cycle includes a dormant stage that can last for several decades. By awakening dormant stages through resurrection biology, scientists can quantify genetic changes at several time points in the past and observe evolution as it happens in nature.
In the study, we were able to hatch dormant eggs that span two decades and to sequence the genome of 36 resurrected waterfleas from a fish-farming pond. During the two decades, the waterflea population experienced a transition from no predation from fish to high fish predation, and back to lower levels of predation.
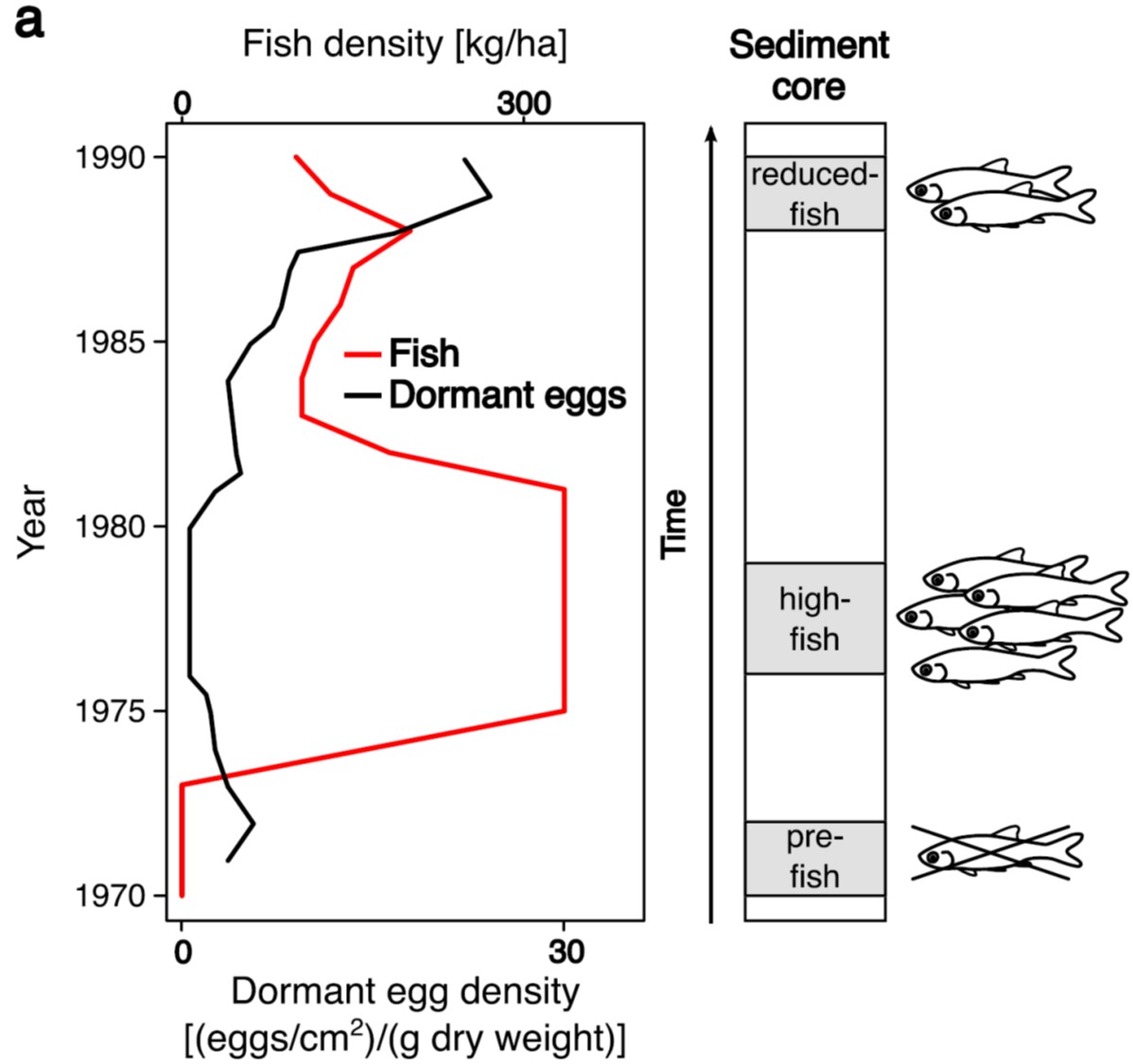
We were able to uniquely match changes in predation pressure on the waterfleas with changes in their DNA over time. We thus managed to link specific changes in the environment of water fleas with the evolution occurring in their genomes.
We found that the required DNA variation to kick-start an evolutionary change that spreads through an entire population did not require more than five founding individuals from the regional set of waterflea populations.
This surprising result suggests that animal species such as waterfleas have a high capacity for adaptive evolution, thanks to the fact that genetic variation is maintained at the landscape level – an important lesson for conservation biology.
Lead researcher, Dr Anurag Chaturvedi, currently at the University of Birmingham’s School of Biosciences, explained: “This type of research will be invaluable for understanding the potential impacts of future environmental changes on animal populations.”.
Further reading